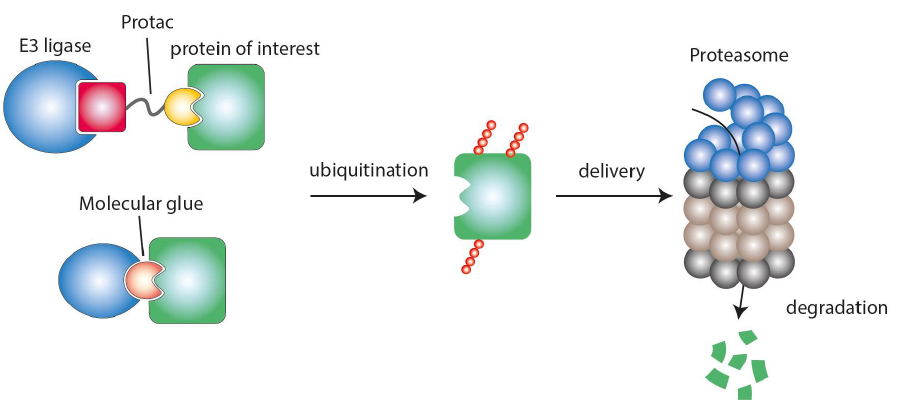
The aim of the platform is to make the drug class of proximity inducing drugs (PiDs) usable for the treatment of immune-mediated diseases. PiDs are bifunctional molecules which, by transient binding to two target structures, bring them into spatial proximity to each other and thereby trigger a biological effect. In PiDs in the narrow sense, one of the two structures is a ubiquitin E3 ligase, and the other is the target protein, which is tagged by this E3 ligase for degradation in the proteasome. These PiDs thus lead to the complete loss of the target protein with all its functions, e.g. catalytic, structural or regulatory, which distinguishes PiDs from classical inhibitors.
80% of the human proteome is considered »undruggable«
Despite a large arsenal of available drug classes, approximately 80% of the human proteome is considered therapeutically undruggable or difficult to access. An active site is not required for PiDs to act - making it possible to address disease-relevant target proteins previously considered undruggable.
PiDs, specifically PROTACs (proteolysis-targeting chimeras), are a new but intensively researched class of molecules. The most advanced compounds of this class are currently in the early stages of clinical development, mainly for oncological diseases.
Characterization and optimization of PiDs
The work within the platform is intended to establish the infrastructure, processes and workflows for the design, synthesis, characterization and optimization of PiDs in a cross-institutional collaboration. Two classes of PiDs will be addressed: PROTACS and LYTACs (lysosomal-targeting chimeras). Using one or more selected target proteins as examples, PiDs will be identified, their efficacy tested in models of immuno-inflammatory diseases and their mode of action compared with that of classical inhibitors.
 Fraunhofer Cluster of Excellence Immune-Mediated Diseases
Fraunhofer Cluster of Excellence Immune-Mediated Diseases